A second post in 2 days on mixture modelling? No awards for guessing what type of analysis I’ve been preoccupied with recently!
Today’s post provides an ugly hack to fix a bug in the R flexmix
package for likelihood-based mixture modelling and provides a cautionary
tale about environments. In short, I’ve encountered problems when trying
to predict the cluster membership for out-of-sample data using this
package, and judging from a
couple
of posts
I found online, I’m not the only one.
Simulating data
Ok let’s get to it. I’ll simulate two different datasets:
- A mixture of 2 univariate Gaussians
- A mixture of 2 bivariate Gaussians
The univariate mixture is shown below with means 120 and 20, and both have a standard deviations of 10.
library(flexmix)
library(tidyverse)
N <- 1000
set.seed(17)
univariate <- rnorm(N, c(120, 20), c(10, 10))
ggplot(as.data.frame(univariate), aes(x=univariate)) +
geom_histogram(colour='black', fill='white') +
theme_bw() +
labs(x="Y", y="Count", title="Univariate mixture of 2 Gaussians")
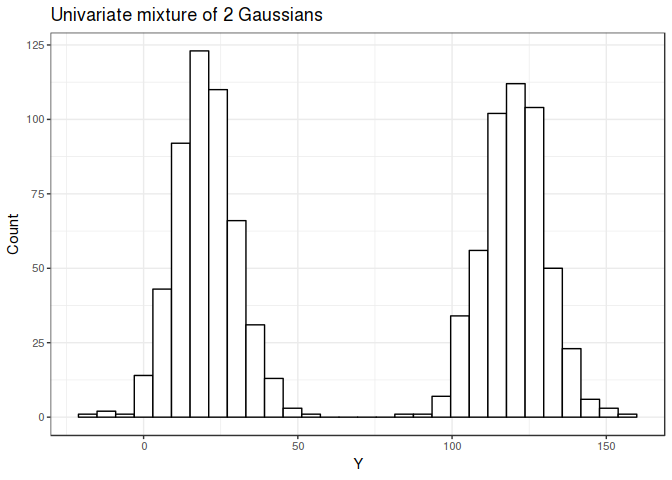
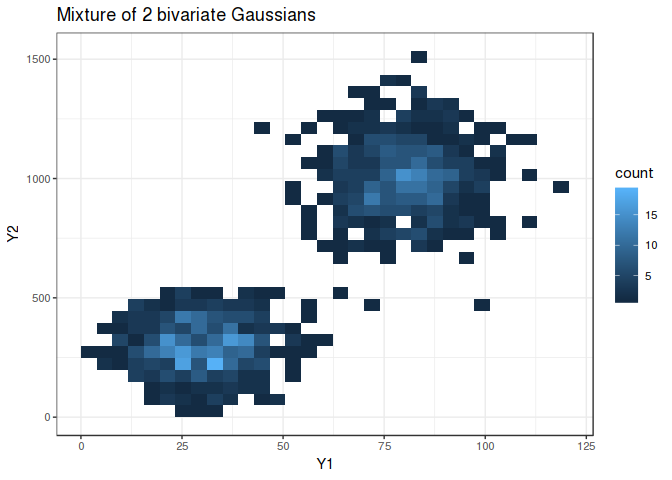
The bivariate mixture is shown below with the two groups cleanly separated (if only real world data looked like this…)
set.seed(17)
bivariate <- matrix(c(rnorm(N, c(80, 1000), c(10, 150)),
rnorm(N, c(30, 300), c(10, 100))),
ncol=2, byrow=TRUE)
colnames(bivariate) <- c('Y1', 'Y2')
as.data.frame(bivariate) %>%
ggplot(aes(x=Y1, y=Y2)) +
geom_bin2d() +
theme_bw() +
labs(title="Mixture of 2 bivariate Gaussians")
Model fitting
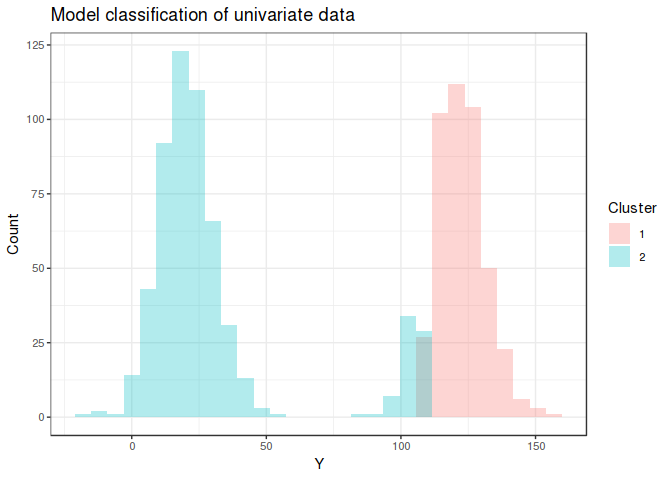
Using flexmix to fit a model with *K* = 2 for the first mixture is
mostly able to distinguish the 2 underlying distributions, although I’m
surprised values > 100 are included in the first group.
set.seed(17)
model_uni <- flexmix(univariate ~ 1, k = 2, control=list(iter=100),
model=FLXMRglm(family="gaussian"))
model_uni
##
## Call:
## flexmix(formula = univariate ~ 1, k = 2, model = FLXMRglm(family = "gaussian"),
## control = list(iter = 100))
##
## Cluster sizes:
## 1 2
## 428 572
##
## convergence after 3 iterations
data.frame(univariate, cluster=model_uni@cluster) %>%
ggplot(aes(x=univariate, fill=as.factor(cluster))) +
geom_histogram(alpha=0.3, position='identity') +
scale_fill_discrete("Cluster") +
theme_bw() +
labs(x="Y", y="Count", title="Model classification of univariate data")
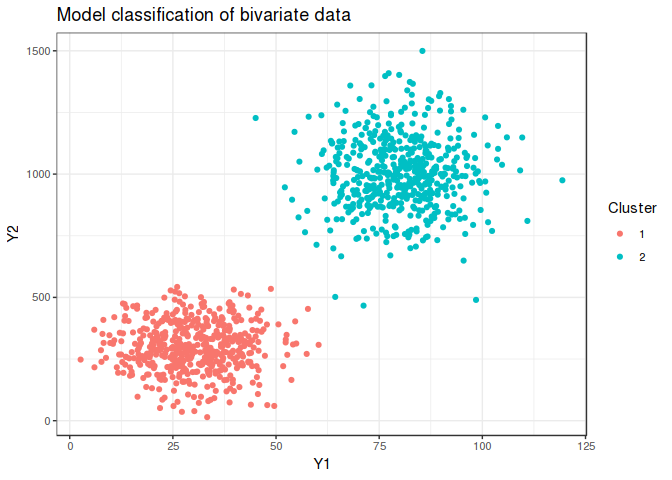
Modelling is completely successful for the somewhat easier bivariate case owing to the lack of overlap.
set.seed(17)
model_bi <- flexmix(bivariate ~ 1, k = 2, model=FLXMCmvnorm(), control=list(iter=100))
model_bi
##
## Call:
## flexmix(formula = bivariate ~ 1, k = 2, model = FLXMCmvnorm(),
## control = list(iter = 100))
##
## Cluster sizes:
## 1 2
## 500 500
##
## convergence after 12 iterations
data.frame(bivariate, cluster=model_bi@cluster) %>%
ggplot(aes(x=Y1, y=Y2, colour=as.factor(cluster))) +
geom_point() +
scale_colour_discrete("Cluster") +
theme_bw() +
labs(x="Y1", y="Y2", title="Model classification of bivariate data")
I’ll now save both these models to be used for future work in classifying as yet unseen data.
saveRDS(model_uni, "model_uni.rds")
saveRDS(model_bi, "model_bi.rds")
Classification
To simulate applying these models for new cases I’ll clear my R environment and load the models, as if we are in a production setting in a data science pipeline.
rm(list=ls())
model_uni <- readRDS("model_uni.rds")
model_bi <- readRDS("model_bi.rds")
Univariate
We’ve now received some new data, let’s see if we can predict which clusters they have arisen from. Note I’ll be attempting to predict the discrete cluster label rather than the posterior probability of each cluster, which seems relatively poorly calibrated from my initial experiments.
We have 3 samples of Y, the univariate variable, which are clear cluster 1, clear cluster 2, and a halfway point.
ndata_uni <- c(150, 20, 70)
But plugging this straight into flexmix::clusters doesn’t work as
expected, seemingly because I am passing in the data as a numeric
vector.
clusters(model_uni, newdata=ndata_uni)
## Error in (function (classes, fdef, mtable) : unable to find an inherited method for function 'posterior' for signature '"flexmix", "numeric"'
Let’s try wrapping it as a data frame.
ndata_uni_df <- as.data.frame(ndata_uni)
ndata_uni_df
## ndata_uni
## 1 150
## 2 20
## 3 70
Ah that’s odd… it complains that it can’t find the variable name of the original training data…
clusters(model_uni, ndata_uni_df)
## Error in eval(predvars, data, env): object 'univariate' not found
Let’s try using this name for the data frame column.
ndata_uni_df2 <- data.frame(univariate=ndata_uni)
ndata_uni_df2
## univariate
## 1 150
## 2 20
## 3 70
And there we go, and it gets the predictions correct and predicts the midway point as cluster 2, which we’d expect from the histogram.
clusters(model_uni, ndata_uni_df2)
## [1] 1 2 2
NB: what I mean about “poorly calibrated posteriors” is shown below, where there is a <1% posterior difference for all 3 points.
posterior(model_uni, ndata_uni_df2)
## [,1] [,2]
## [1,] 0.5042937 0.4957063
## [2,] 0.4908194 0.5091806
## [3,] 0.4959340 0.5040660
Bivariate
Now let’s do the same for the bivariate case, these new measurements should be assigned to cluster 2 which had means of 80 and 1000.
ndata_bi <- c(80, 1000)
Again, we get the error about not being able to pass vectors into
clusters.
clusters(model_bi, newdata=ndata_bi)
## Error in (function (classes, fdef, mtable) : unable to find an inherited method for function 'posterior' for signature '"flexmix", "numeric"'
Again trying to pass the values in as a data frame results in an error
message that it cannot find the bivariate variable.
ndata_bi_df <- data.frame(y1=80, y2=1000)
clusters(model_bi, newdata=ndata_bi_df)
## Error in eval(predvars, data, env): object 'bivariate' not found
The solution for the univariate case was to pass in a data frame where the column had the name of the original matrix, but obviously this won’t work here. Perhaps calling this data frame the same as the original training data will do the trick?
Nope, this still fails.
bivariate <- data.frame(Y1=80, Y2=1000)
clusters(model_bi, newdata=bivariate)
## Error in model.frame.default(model@terms, data = data, na.action = NULL, : invalid type (list) for variable 'bivariate'
Solution
After a while of tearing my hair out, I managed to fix this with a particularly beautiful hack, shown below.
Essentially, the code needs a variable with the same name and data type
as the original training data (bivariate in this case and a matrix).
It uses eval to run the prediction, which looks for this variable in
the current environment, hence the use of the <-- operator, which
assigns this name in the global environment.
The other point to note is that the newdata keyword is expecting a
data frame, not a matrix, so we must pass it such. However, it never
actually uses the passed in value, so as long as this is a data frame
with the same number of rows as the actual data it doesn’t matter what
the values are.
predict_cluster <- function(y1, y2) {
bivariate <<- matrix(c(y1, y2), ncol=2)
clusters(model_bi, newdata=data.frame(dummy=rep(1, length(y1))))
}
This function now correctly classifies the means of the two distributions.
predict_cluster(80, 1000)
## [1] 2
predict_cluster(30, 300)
## [1] 1
And by setting the dummy data frame length, it can be used for arbitrary length input.
predict_cluster(c(80, 30, 50), c(1000, 300, 500))
## [1] 2 1 1
That’s it for today, the lesson being take extreme care when using
eval in your code and to setup a high coverage testing suite.
Development of the package looks to have gone stale, and the source code
isn’t on GitHub else I would have submitted this fix as a PR. Please let
me know if you’ve also encountered this issue and this post has helped!