I’ve finally managed to combine my two main hobbies of data science and running into one project: a Shiny web-app that allows you to explore your Strava fitness data, as well as providing a local database of all your activities that you can analyse with R.
My initial motivation was that I wanted quick access to certain visualisations and metrics that either Strava doesn’t provide, or are awkward to get so I wrote a basic app for my own use.
CONTINUE READING
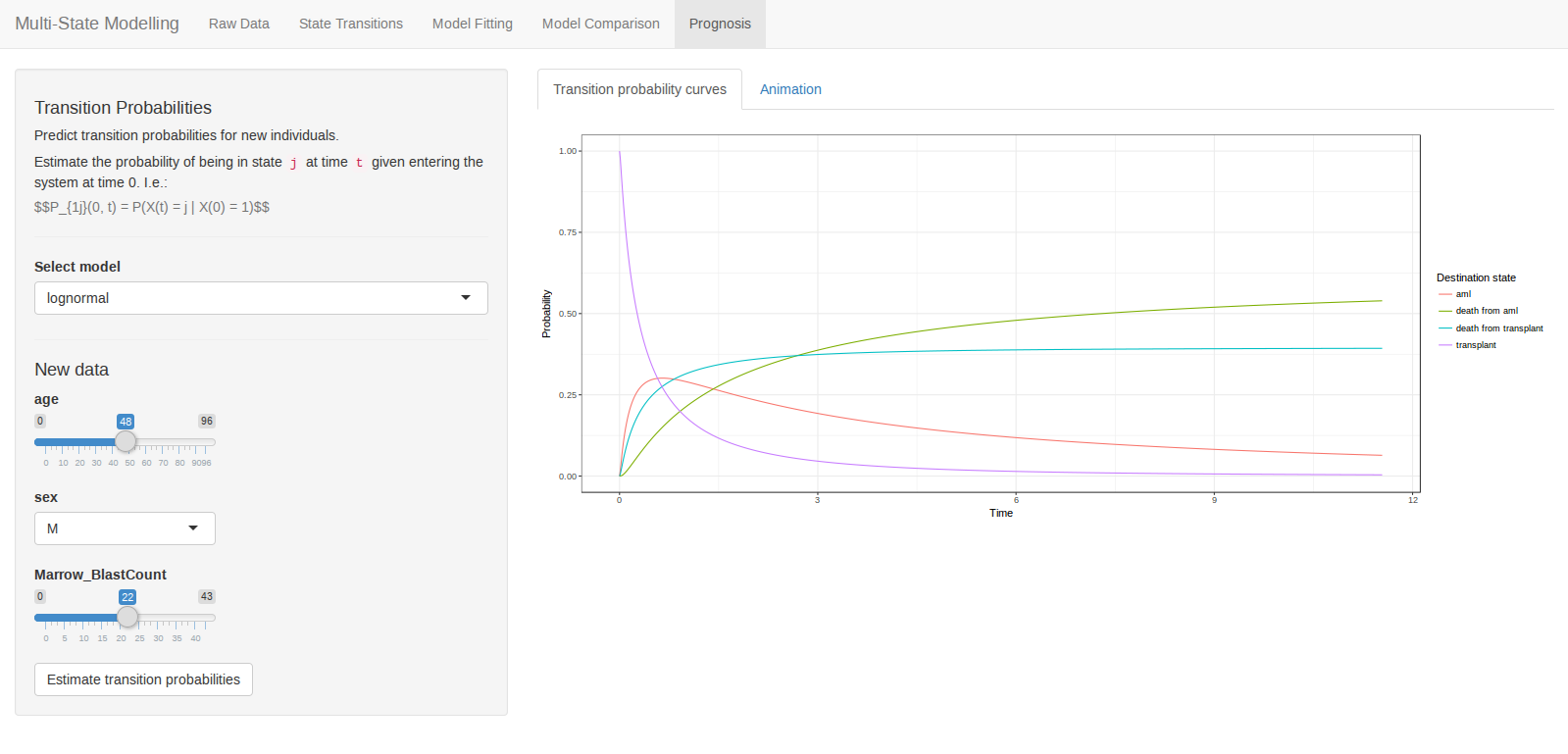
Introduction Deep learning has attracted considerable attention for its near-human ability in a variety of complex problems such as image recognition, playing games, and recently conversational AI through large language models. Each of these applications requires unimaginable volumes of data and computational resources beyond the reach of all but the richest companies. This resource hungry nature, coupled with the huge hype that accompanies any deep-learning application, makes it challenging to gain a realistic assessment of their real-world potential for less demanding use-cases, such as scientific time-series modelling.
CONTINUE READING
Just a quick update, more to test that the website infrastructure is still running than anything else, as it’s been 4 years since my last post. I ran a workshop (slides here) at the University’s Research Coding Club on speeding up data analysis in R last month that might be useful for anyone who stumbles across this page in the future.
The Research Coding Club is an informal collective of people from across the entire University who write software to aid their research.
CONTINUE READING
In the previous entry of what has evidently become a series on modelling binary mixtures with Dirichlet Processes (part 1 discussed using pymc3 and part 2 detailed writing custom Gibbs samplers), I ended by stating that I’d like to look into writing a Gibbs sampler using the stick-breaking formulation of the Dirichlet Process, in contrast to the Chinese Restaurant Process (CRP) version I’d just implemented.
Actually coding this up this was rather straight forward and took less time than I expected, but I found the differences and similarities between these two same ways of expressing the same mathematical model interesting enough for a post of its own.
CONTINUE READING
Back at the start of the year (which really doesn’t seem like that long a time ago) I was looking at using Dirichlet Processes to cluster binary data using PyMC3. I was unable to get the PyMC3 mixture model API working using the general purpose Gibbs Sampler, but after some tweaking of a custom likelihood function I got something reasonable-looking working using Variational Inference (VI). While this was still useful for exploratory analysis purposes, I’d prefer to use MCMC sampling so that I have more confidence in the groupings (since VI only approximates the posterior) in case I wanted to use these groups to generate further research questions.
CONTINUE READING